Research
We are interested in the neurobiological foundations underlying the causes, mechanisms, and trajectories of psychiatric and neurological disorders. Our work is deeply rooted in development. We take advantage of human pluripotent stem cells to reconstitute and understand human brain development. By developing new high-throughput in vitro model systems and employing single-cell omics approaches, we seek to understand how neural tissue is established during fetal development, how neurons form interconnected networks, and how they vary among individuals and in psychiatric and neurological disorders. We are committed to uncovering the regulatory networks that control stem cell differentiation and developing novel methods to increase the precision and safety of stem cell-based therapies. In addition to the in vitro model systems, we develop and use new experimentally tractable animal models of human disease to interrogate disease mechanisms and discover new therapeutic opportunities.
Yang Laboratory
Nan Yang, Ph.D.
Associate Professor, Neuroscience
Location
Lab: Ichan 13-52
Office: Icahn 13-20D
Phone
Office: 212-659-8288
Lab: 212-559-1379
Email: nan.yang1@mssm.edu
Projects
Brain development and Reprogram
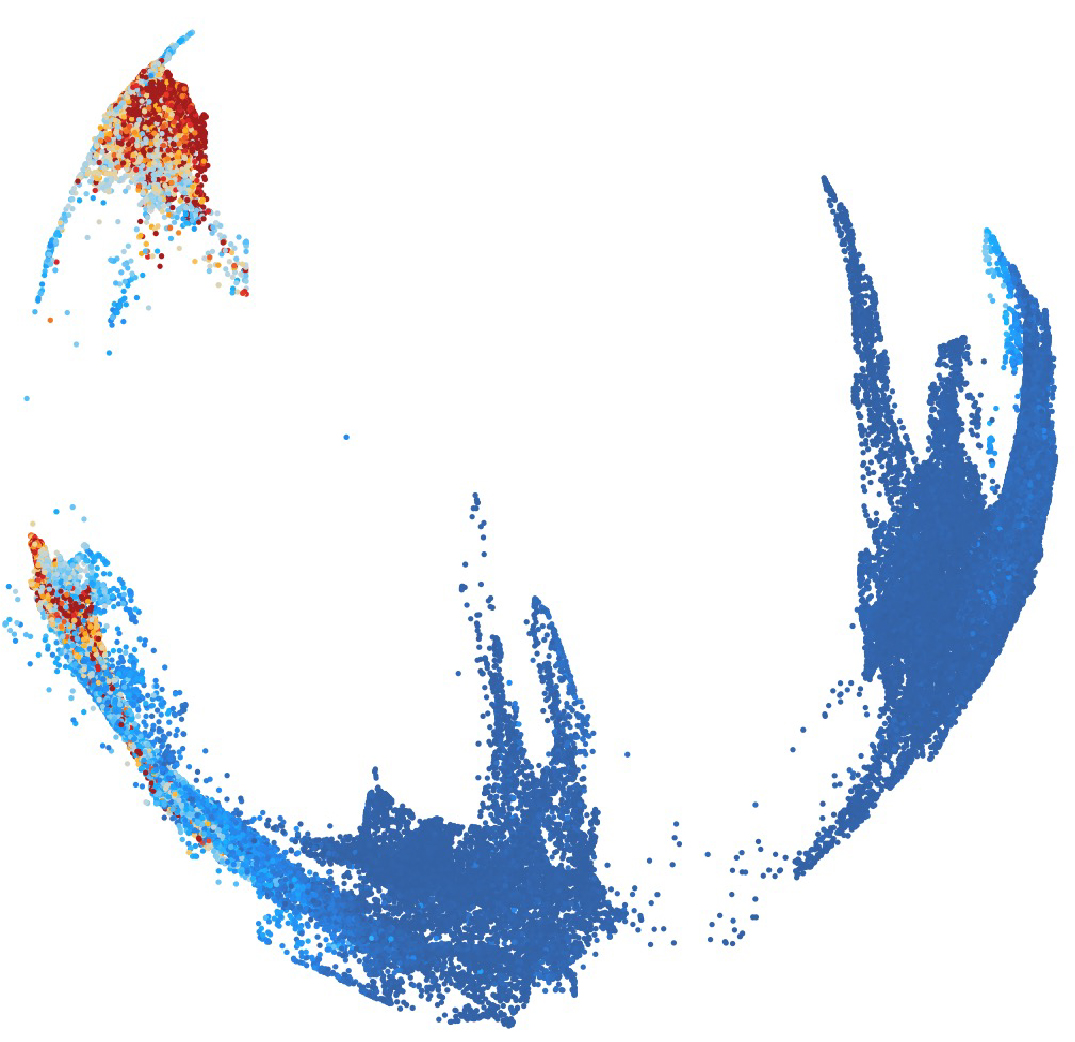 The formation of the central nervous system is a remarkable process that starts with the generation of and interaction between diverse cell types. Using single-cell profiling that captures cell identity and cell states within a complex tissue, our work seeks to unveil the series of rules underlying the extraordinary cellular diversity and human brain organization. By applying our knowledge of development, we recreate 2D and 3D in vitro systems to recapitulate key events during human brain development. These systems promise to uncover the principles of cell diversification and create a platform for stem cell-based disease investigation.
The formation of the central nervous system is a remarkable process that starts with the generation of and interaction between diverse cell types. Using single-cell profiling that captures cell identity and cell states within a complex tissue, our work seeks to unveil the series of rules underlying the extraordinary cellular diversity and human brain organization. By applying our knowledge of development, we recreate 2D and 3D in vitro systems to recapitulate key events during human brain development. These systems promise to uncover the principles of cell diversification and create a platform for stem cell-based disease investigation.
Neurodevelopmental Disorders
Our team aims to translate genetic discoveries from human studies into actionable biological hypotheses and provide new insights into the causal chain of events that result in neurological and psychiatric disease conditions. Together with the Seaver Autism Center at Mount Sinai, we apply human stem cell-based and mouse model systems and use multidisciplinary approaches, including CRISPR gene editing, electrophysiology, single-cell sequencing, and high-resolution imaging, to learn how the disease-associated genetic variation impairs neuronal properties and circuit activities. We also employ brain organoids combined with live-imaging and single-cell profiling to study the early molecular and cellular events, such as cell proliferation, migration, and differentiation impaired by disease-risk variants identified in recent large-scale genomic studies.
Neuroepigenetics and diseases
Disorders of the brain affect nearly one-fifth of the world’s population. Although genome-wide association studies (GWAS) have identified many genetic variants strongly linked to neurological and psychiatric disorders, functional interpretation is challenging. Most heritable common disease risk distributes to non-coding regions and appears to be particularly enriched in enhancers (classically defined as cis-acting DNA sequences that can increase the transcription of genes) that are specific to disease-relevant cell types. However, only a handful of the enhancers nominated via biochemical annotations have been validated and confidently linked to their target genes. We use innovative multidisciplinary approaches built upon stem cell technology to unravel the functions of non-coding sequences in disease-relevant neuronal types. This work promises an exploration of the biological functions of non-coding regions containing disease risk variants and helps bridge the gap between human genetics and experimental biology.
Publications
2022
Mapping cis-regulatory elements in human neurons links psychiatric disease heritability and activity-regulated transcriptional programs
Sanchez-Priego C, Hu R, Boshans LL, Lalli M, Janas JA, Williams SE, Dong Z, Yang N, Cell Reports. 2022
Generation of Functional Human Oligodendrocytes from Dermal Fibroblasts by Direct Lineage Conversion
2021
Direct Differentiation of Functional Neurons from Human Pluripotent Stem Cells (hPSCs).
Efficient generation of dopaminergic induced neuronal cells with midbrain characteristics.
2019
Oligodendrocyte Death in Pelizaeus-Merzbacher Disease Is Rescued by Iron Chelation.
Cell Stem Cell. 2019 Oct 3;25(4):531-541.e6.
Neuroligin-4 Regulates Excitatory Synaptic Transmission in Human Neurons.
2018
2017
A Peek into Parkinson’s Disease Progression through Human Dopamine Neurons in a Dish.
μNeurocircuitry: Establishing in vitro models of neurocircuits with human neurons.
Generation of pure GABAergic neurons by transcription factor programming.
Earlier than 2015
Transdifferentiation of mouse fibroblasts and hepatocytes to functional neurons.
Harnessing the stem cell potential: a case for neural stem cell therapy.
Rapid single-step induction of functional neurons from human pluripotent stem cells.
Generation of oligodendroglial cells by direct lineage conversion.
Induced neuronal cells: how to make and define a neuron.
Direct lineage conversion of terminally differentiated hepatocytes to functional neurons.
Induction of human neuronal cells by defined transcription factors.
Genomic selection identifies vertebrate transcription factor Fezf2 binding sites and target genes.
A systematic approach to identify functional motifs within vertebrate developmental enhancers.
A subunit of the mediator complex regulates vertebrate neuronal development.
Meet the Team

Aleta Murphy
Graduate student
Aleta is a Ph.D. student in Neuroscience. She is interested in using human stem cell technologies to investigate the impact of autism spectrum disorders (ASD) -associated genetic variations on neural development and function. Outside the lab, Aleta enjoys reading mystery novels, picnicking in Central Park, and consuming lots of bubble tea.

Linda Lee
Postdoc
Linda is a postdoctoral fellow in the department of neuroscience. Her project involves using iPS cell-derived 3D cerebral organoids to investigate the role of NF1 on cellular and molecular mechanisms of early brain development. In her free time, she enjoys dancing, biking, traveling, and making ice cream.

Ratchell Sadovnik
Graduate student
Ratchell is a Ph.D. student in Biomedical Sciences, with a focus on Development, Regeneration, and Stem Cells. Her research uses an organoid model to study how autism-related mutations affect neurodevelopment. In her free time, Ratchell enjoys reading, playing with her dogs, traveling, and exploring the city.

Sarah Williams
Graduate student
Sarah is a Ph.D. student in the department of neuroscience, co-mentored by Dr. Kristen Brennand of Yale University. She is interested in using human stem cell models to study genetic risk for psychiatric disorders, with an emphasis on cell-type-specific effects, non-coding variants, and multiplexing strategies. Outside of lab, Sarah enjoys traveling, photography, and exploring all of NYC’s museums, live shows, and antique shops.

Yvette Somersel
Undergraduate student
Yvette is an undergraduate student at NYU majoring in neuroscience. She has been participating in research with other team members since 2020.
Lab Alumni
Alida Pahlevan (Northeastern University): Summer Undergraduate (2022)
Jacqueline (Jackie) Cho (Brown University): Summer Undergraduate (2022)
Gizem Inak Girrbach (Nuvisan ICB in Berlin, Associate Scientist): postdoc (2020-2021)
Carlos Sanchez-Priego (Genentech Inc.): graduate student (2017 – 2021)
Stephanie Rogers (University of Cambridge): research assistant (2018 – 2019)

Henry Kim
Undergraduate student
Henry is a Research Associate working in both Dr. Nan Yang and Dr. Zhenyu Yue’s labs. His current project works to elucidate novel adaptors in the selective degradation of the endoplasmic reticulum. When he is not working, he likes to spend time with his dogs and listen to music and podcasts.

Mark Youssef
Research Associate
Mark joined the lab in June 2021 as part of the Summer Undergraduate Research Program and then came back as a research assistant in 2022 after graduating from Rutgers University for his gap year before he applies to MD-PhD Programs.

Ruiqi (Ritchi) Hu
Postdoctoral Fellow
Ritchi studies molecular mechanisms of human neurogenesis and develops innovative human models for neurological and psychiatric disease investigation. She aims to use high-throughput methods to examine autism-linked genetic variants and was awarded the Seaver Center for Autism Fellowship. Ritchi enjoys her leisure time reading, sports, music, and crafts.

Xiaoting Zhou
Graduate student
Gallery and News
Contact us
Whether you’re looking to collaborate or want to become an ambitious scientist and become part of our team, the Yang lab is eager to connect. Please email us (nan.yang1@mssm.edu)
Postdoctoral Fellows
Are you a passionate scientist with a background in Neuroscience, Molecular Biology, or Cellular Biology? We are currently seeking highly motivated Postdoctoral Fellows to join our lab and contribute to our exciting research projects. We are looking for individuals who are goal-driven, curious, and supportive team players with a critical mindset.
At our lab, we value diverse perspectives and encourage our team members to bring their unique skillset to the table. Our goal is to help you reach your full potential and develop your vision as an independent scientist, whether in or out of academia. Most importantly, we want you to have fun and find meaning in the work you do with us.
If you are interested in becoming a part of our team, please contact us. We can’t wait to hear from you!
Graduate Students
We warmly welcome highly motivated graduate students to come and rotate with us! We are seeking curious, dedicated individuals who are excited about our research and ready for a challenge. As a member of our team, you will have the opportunity to develop your own projects and receive comprehensive mentorship every step of the way. From conceiving and designing the study, to data collection and analysis, and all the way to publication and career development, we will be there to support you.
Current students can contact Nan directly to discuss rotation opportunities, while prospective students can refer to the ISMMS Graduate Admissions resources to apply. We are committed to helping you achieve your goals and realize your full potential as a researcher. So, if you are passionate about science and eager to learn, come and join us!





