Research
Zou lab is interested in molecular mechanisms of how neurons are born, how they extend axons, and why they fail to regenerate after CNS injury. We combine in vivo axon injury models, 3D organoid cultures, molecular biology, and various imaging and tissue engineering techniques to study novel signaling pathways and epigenetic mechanisms that promote neurogenesis and axonogenesis. Such knowledge is crucial for targeting molecules for effective CNS regeneration. Similar mechanisms that govern neural stem cell biology also have relevance in the malignant potency of tumor stem cells in glioblastoma, the most malignant type of brain tumor. The lab is dissecting the governing factors of glioma stem cell dormancy and tumor microenvironment, using both biomimetic 3D vascular glioma model and patient-derived xenograft model.

Projects
Background
CNS Diseases
- In cases of brain or spinal cord injury, there is limited neuronal or axonal regeneration, due to both neuronal intrinsic and extrinsic barriers. The first research area of our laboratory is molecular mechanisms that promote axon growth potential in adult neurons. Neuroinflammation and glial reaction also heavily influence the functional outcome of neural repair, and our laboratory is actively investigating novel signaling pathways that govern glial response and neuroimmunity after CNS injury.
- The second research area is to dissect how neural stem cells respond to injury signals in order to develop new strategies for neural repair.
- Glioma stem cells share many similar molecular characteristics as neural stem cells. The third research area is to understand how tumor stem cells acquire dormancy and what are the niche factors in tumor microenvironment that are important for glioma quiescence.
Project 1: Molecular mechanisms of axon regeneration
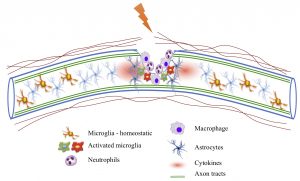
- Transcriptional regulation of axon growth gene program.
How to promote axon regeneration remains an important unsolved question in neurobiology. Failure of CNS axon regeneration has been attributed to an inhibitory environment and an age-dependent decline of intrinsic axon growth potential. Our laboratory has been actively studying the close collaboration between regeneration-associated transcription factors and epigenetic machinery in order to turn on the pro-growth gene program in neurons. We combine in vivo mouse spinal cord injury models, neuronal cultures, molecular and epigenetic techniques to study chromatin landscapes and transcriptional mechanisms of regeneration genes.
- Neuroinflammation and glial reactivity after CNS injury.
A hostile injury environment constitutes an extrinsic barrier for successful axon regeneration. Our laboratory combines cell type specific techniques, genome-wide transcriptomic analysis, and mouse genetic approaches to study signaling pathways and epigenetic mechanisms that underlie how microglia and astrocytes interact with one another during recovery phases after spinal cord injury.
Project 2: Neural stem cells plasticity for neural repair after CNS injury
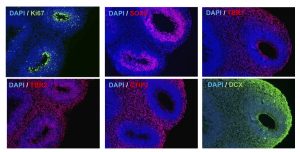
Neural stem cells play an important role in modulating injury response after CNS injury. Our laboratory investigates molecular players that mediate injury response of neural stem cells during neurodevelopment and after CNS injury. We employ a variety of tools that include mouse genetics, stem cell cultures, 3D organoids, stroke models, imaging, and tissue-engineering techniques.
Project 3: Glioma dormancy, stem cell niche and tumor microenvironment
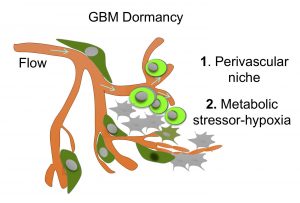 Malignant brain tumors frequently recur after therapy. Dormant tumor stem cells constitute potential sources for tumor recurrence. Our laboratory is teaming up with bioengineers to develop a novel biomimetic 3D glioma vascular model to dissect governing factors of glioma quiescence.
Malignant brain tumors frequently recur after therapy. Dormant tumor stem cells constitute potential sources for tumor recurrence. Our laboratory is teaming up with bioengineers to develop a novel biomimetic 3D glioma vascular model to dissect governing factors of glioma quiescence.
Featured
Meet the Team