Research

In my laboratory, my team and I study how environmental signals and insults are interpreted by our cells. Our mechanism of focus is epigenetics. Epigenetics refers to the functionally relevant modifications to the genome that do not involve a change in the DNA sequence. Epigenetics is the bridge connecting the environment and genetics, regulating gene expression and cellular response to environmental cues throughout our lifespan. To better understand the role of environmental factors in brain function and dysfunction, we investigate epigenetics, and in particular, DNA methylation patterns on a genome scale. We are interested in how physical and/or psychosocial stress can induce epigenetic abnormalities that later lead to neuropathological and psychiatric symptoms. Physical stressors like traumatic brain injury (TBI) can enhance or contribute to these abnormalities, and psychosocial stressors often precipitate psychopathology and heightened suicide risk. Applying both genomics and computational approaches, we undertake human studies to identify genes and genetic pathways that undergo sustained transcriptional regulatory changes in response to such environmental insults.
Connect With Us
Neurobiology of Suicide
According to the CDC, suicide is the 10th leading cause of death in the US. The Veteran population suffers from disproportionately high suicide rates. Understanding the molecular and cellular mechanisms underlying suicide risk is the primary research focus of the lab addressing a critical public health concern. We tackle this in a multi-pronged fashion, undertaking clinical, human postmortem, and large-scale data analytic strategies:
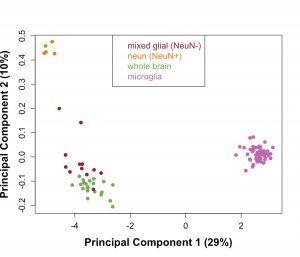
- The clinical studies involve investigation of genome-scale epigenetics (specifically DNA methylation), transcriptional (RNA-seq), and inflammatory signatures in suicidal veterans. Through longitudinal studies, we are characterizing the dynamics of peripheral biomarkers associated with suicide risk to identify objective measures that we can use in clinical settings for suicide risk prediction and to determine the efficacy of treatment course and response.
- The clinical suicide research studies are also complemented by basic neurobiological studies in the brain using postmortem human specimens from suicide decedents, vital to our understanding of the neuropathology of suicide. Our lab is focused on delineating the DNA methylation and transcriptional changes that occur in the brains of suicide decedents in a cell-specific manner to identify aberrant transcriptional regulation unique to suicide neuropathology.
- Through our suicide research as part of the Million Veteran Project (MVP), we are applying big data analytic approaches to identify suicidal subtypes and genetic risk factors associated with suicide risk and behavior.
Blast, TBI & Related Sequelae
With the high incidence of suicide amongst individuals with TBI, we have developed a complementary research program investigating the molecular mechanisms of TBI associated blast injuries. Injuries from exposure to explosive blasts rose dramatically during Operation Iraqi Freedom and Operation Enduring Freedom (OIF, OEF) due to the increased use of improvised explosive devices (IEDs) in military settings, as well as civilian populations through acts of terrorism, which have motivated investigations into blast-related neurotrauma. Despite these increased occurrences, our understanding of blast effects and the mechanisms behind subsequent injury is limited. In recent years, investigations of the blast effect in tactical training settings have begun to address this gap in knowledge. These are primarily studies of breachers, a unique population of military and law enforcement personnel who are, by occupational definition, typically in close proximity to controlled, low-level blast during explosive breaching operations and training, repeatedly exposed to blast overpressure waves. Beyond explosive breaching, military and law enforcement personnel are also repeatedly exposed to blast through indirect fire (artillery, mortar), and antiarmor weapon operations. These exposures have been associated with self-reported concussion-like symptomology, transient decrements in performance, physiological changes including elevations in blood-based neurotrauma biomarkers, and effects on healthcare utilization . Of increasing concern are the evolving chronic neurological and psychological symptoms, including serious and often comorbid disorders such as chronic headaches, cognitive impairment, PTSD, depression, sleep disturbances, and elevated risk for suicide. In our Veterans such evolving clinical symptoms associated with blast exposures and mTBI impacts reintegration difficulties and overall functional disability overtime.
Human Studies of Effects of Repeated Blast

Exposure to blast during breaching
- Research in the lab is focused on identifying transcriptional regulatory changes associated with blast exposure. Leveraging our strong partnership with the VA and DoD, we are investigating repeated exposures to blast in a real-world operational setting to identify DNA methylation signatures of blast that track with associated chronic symptoms both in Service Members and Veterans. We use genomics and machine learning approaches to develop methylation risk scores of cumulative blast and associated symptoms, allowing us to potentially identify those who may be at risk for developing blast-related outcomes.
Clinical
Suicide is the tenth leading cause of death in the US. For every suicide death, there are 25 suicide attempts, according to the American Foundation for Suicide Prevention. Rates of suicide are rising, with higher suicide rates in the Veteran population than the civilian. As part of the clinical efforts in my laboratory at Icahn School of Medicine at Mount Sinai and the Bronx VA (James J. Peters VA Medical Center, JJPVAMC), we are involved in development of innovative suicide treatment interventions that target at-risk Veterans towards the goal of suicide prevention.
Resilience and Wellness Center

The Resilience and Wellness Center (RWC) seeks to create a holistic program dedicated to maximizing the overall wellbeing of our Veterans. Our Veterans require a complete and multi-faceted approach for treatment and services. Our James J. Peters (JJP) Resilience and Wellness Center is at the forefront of developing and testing novel treatment, prevention, and rehabilitation approaches for “high-risk” and “at-risk” Veterans. Complementary and Integrative Health (CIH) interventions are promising therapies in suicide prevention, and in promoting patient engagement. CIH interventions may play a critical role in transitioning veterans into mental health care who do not consistently engage in mental health services, often citing stigma as a barrier.
Resiliency is cultivated through services, programs, and community gatherings designed to promote healing and integrative wellness, provide education, and  bring people together in meaningful dialogue and activity. The mission of the Resilience and Wellness Center is to combat the high rates of death by suicide in Veterans through strong treatment and programmatic components that include physical activity, music and dance, diet and nutrition, horticultural therapy, sleep, spirituality, enhanced peer support and skills-based education classes on stress management and mindfulness. Veterans are referred to the program through hospital-wide consultations from a JJP physician or practitioner.
bring people together in meaningful dialogue and activity. The mission of the Resilience and Wellness Center is to combat the high rates of death by suicide in Veterans through strong treatment and programmatic components that include physical activity, music and dance, diet and nutrition, horticultural therapy, sleep, spirituality, enhanced peer support and skills-based education classes on stress management and mindfulness. Veterans are referred to the program through hospital-wide consultations from a JJP physician or practitioner.
- Program evaluation of RWC, showed that an in-person program with an intensive complement of CIH interventions resulted in significant improvement in suicidal ideation and associated risk factors i.e., depression severity, hopelessness symptoms, with high veteran engagement. (Vitale A, Byma L, Sun S, et al. Effectiveness of Complementary and Integrative Approaches in Promoting Engagement and Overall Wellness Toward Suicide Prevention in Veterans. Journal of Alternative and Complementary Medicine. 2020.)

- In a pilot program evaluation that was driven by the COVID-19pandemic, we set out to see whether the virtual delivery of this multimodal CIH intervention was similarly effective. Indeed, virtual delivery of the RWC CIH interventions resulted in significant reduction in depression and PTSD severity, with those Veterans having prior history of suicidal ideation or attempt showing significant improvements in stress coping skills. This pilot study suggests that virtual delivery of CIH interventions may be impactful in improving overall wellness and broaden access to care for at-risk populations. (Ganesh N, Byma L, Sun S, et al. Pilot Study of Virtual Delivery of Multimodal Complementary Integrated Health Intervention for Suicide Prevention. 2021 IASR/AFSP International Summit on Suicide Research. Oct 24-27. Barcelona, Spain)
- Through a community partnership between the Bronx VA and the New York Botanical Garden, NYBG, has been providing a specially designed combination of horticultural therapy and gardening activities to RWC participating Veterans since January 2020. This program—called Therapeutic Horticulture and Rehabilitative Interventions for Veteran Engagement, or THRIVE—was developed through funding support from the Institute of Museum and Library Services. Program evaluation of this horticultural therapeutic intervention amongst Veterans at-risk, with prior history of suicidal ideation or attempt showed significant reductions in stress, depressed mood, and loneliness that were sustained after a single dose of horticultural therapeutic intervention.(Meore A, Sun S, Byma L, et al. Pilot evaluation of horticultural therapy in improving overall wellness in veterans with history of suicidality. Complementary therapies in medicine. 2021:102728.)
Meet the PI

Getting a closer look… Medusa bust at the Basilica Cistern in Istanbul, Turkey
Dr. Haghighi obtained her PhD in Statistical Genetics and Postdoctoral Fellowship in Bioinformatics from Columbia University. She is a Professor at the Icahn School of Medicine at Mount Sinai and a Senior Researcher at the James J. Peters VA Medical Center (JJPVAMC). Her research focuses on understanding how environmental exposures, such as stress and injury, result in molecular changes and disease. To this end, she has spearheaded research studies at the JJPVAMC investigating the effect of repeated blast on inducing aberrant gene regulation. Additionally, she has developed a robust suicide research portfolio examining the neuroinflammatory and epigenetic changes associated with chronic stress and risk of suicide, both in the peripheral and central nervous systems.
Outside of work, Dr. Haghighi enjoys traveling and being a foodie (often at the same time) and keeping up with the latest accessible technology as a blind scientist with Stargardt disease.
Meet the Team




Lab Alumni

Rachel Harris
Alumnus
PsyD candidate at William Paterson University

Sharon Alter
Alumnus
Now with Dr. Szesko at the Bronx VAMC


Natalia Mendelev
Alumnus
Now with Dr. Sealfon at Mount Sinai


Amanda Vitale
Research Coordinator
PhD Student at Montclair State University




